Abstract
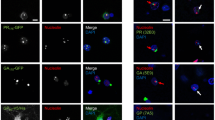
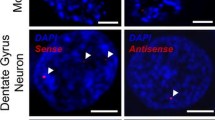
Genetic analysis revealed the hexanucleotide repeat expansion GGGGCC within the regulatory region of the gene C9orf72 as the most common cause of familial amyotrophic lateral sclerosis and the second most common cause of frontotemporal lobar degeneration. Since repeat expansions might cause RNA toxicity via sequestration of RNA-binding proteins, we searched for proteins capable of binding to GGGGCC repeats. In vitro-transcribed biotinylated RNA containing hexanucleotide GGGGCC or, as control, AAAACC repeats were incubated with nuclear protein extracts. Using stringent filtering protocols 20 RNA-binding proteins with a variety of different functions in RNA metabolism, translation and transport were identified. A subset of these proteins was further investigated by immunohistochemistry in human autopsy brains. This revealed that hnRNP A3 formed neuronal cytoplasmic and intranuclear inclusions in the hippocampus of patients with C9orf72 repeat extensions. Confocal microcopy showed that these inclusions belong to the group of the so far enigmatic p62-positive/TDP-43 negative inclusions characteristically seen in autopsy cases of diseased C9orf72 repeat expansion carriers. Thus, we have identified one protein component of these pathognomonic inclusions.




Similar content being viewed by others
References
Al-Sarraj S, King A, Troakes C et al (2011) p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol 122(6):691–702
Arai T, Hasegawa M, Akiyama H et al (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351(3):602–611
Baker M, Mackenzie IR, Pickering-Brown SM et al (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442(7105):916–919
Benajiba L, Le Ber I, Camuzat A et al (2009) TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol 65(4):470–473
Brettschneider J, Van Deerlin VM, Robinson JL et al (2012) Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol 123(6):825–839
Collins M, Riascos D, Kovalik T et al (2012) The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients. Acta Neuropathol 124(5):717–732
Cruts M, Gijselinck I, van der Zee J et al (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442(7105):920–924
DeJesus-Hernandez M, Mackenzie IR, Boeve BF et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72(2):245–256
Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11(5):1475–1489
Dormann D, Madl T, Valori CF et al (2012) Arginine methylation next to the PY-NLS modulates transportin binding and nuclear import of FUS. EMBO J 31(22):4258–4275
Elden AC, Kim HJ, Hart MP et al (2010) Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466(7310):1069–1075
Fratta P, Mizielinska S, Nicoll AJ et al (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep 2:1016
Gasser T, Hardy J, Mizuno Y (2011) Milestones in PD genetics. Mov Disord 26(6):1042–1048
Gijselinck I, Van Langenhove T, van der Zee J et al (2012) A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol 11(1):54–65
Gomez-Tortosa E, Gallego J, Guerrero-Lopez R et al (2013) C9ORF72 hexanucleotide expansions of 20–22 repeats are associated with frontotemporal deterioration. Neurology. doi:10.1212/WNL.0b013e31827f08ea
Haass C, Selkoe DJ (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol 8(2):101–112
He Y, Smith R (2009) Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci 66(7):1239–1256
Hilleren PJ, Parker R (2003) Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell 12(6):1453–1465
Keller BA, Volkening K, Droppelmann CA et al (2012) Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol 124(5):733–747
Kwiatkowski TJ Jr, Bosco DA, Leclerc AL et al (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323(5918):1205–1208
Ma AS, Moran-Jones K, Shan J et al (2002) Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J Biol Chem 277(20):18010–18020
Mackenzie IR, Neumann M, Baborie A et al (2011) A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol 122(1):111–113
Maruyama H, Morino H, Ito H et al (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465(7295):223–226
Neumann M, Kwong LK, Lee EB et al (2009) Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol 117(2):137–149
Neumann M, Sampathu DM, Kwong LK et al (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314(5796):130–133
Papadopoulou C, Boukakis G, Ganou V et al (2012) Expression profile and interactions of hnRNP A3 within hnRNP/mRNP complexes in mammals. Arch Biochem Biophys 523(2):151–160
Rademakers R, Neumann M, Mackenzie IR (2012) Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol 8(8):423–434
Ranum LP, Cooper TA (2006) RNA-mediated neuromuscular disorders. Annu Rev Neurosci 29:259–277
Renton AE, Majounie E, Waite A et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72(2):257–268
Rosen DR, Siddique T, Patterson D et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362(6415):59–62
Schreiner B, Westerburg H, Forne I et al (2012) Role of the AAA protease Yme1 in folding of proteins in the intermembrane space of mitochondria. Mol Biol Cell doi. doi:10.1091/mbc.E12-05-0420
Shevchenko A, Chernushevich I, Wilm M, Mann M (2000) De Novo peptide sequencing by nanoelectrospray tandem mass spectrometry using triple quadrupole and quadrupole/time-of-flight instruments. Methods Mol Biol 146:1–16
Sieben A, Van Langenhove T, Engelborghs S et al (2012) The genetics and neuropathology of frontotemporal lobar degeneration. Acta Neuropathol 124(3):353–372
Simon-Sanchez J, Dopper EG, Cohn-Hokke PE et al (2012) The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain 135(Pt 3):723–735
Skibinski G, Parkinson NJ, Brown JM et al (2005) Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 37(8):806–808
Sreedharan J, Blair IP, Tripathi VB et al (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319(5870):1668–1672
Thompson DM, Parker R (2007) Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol 27(1):92–101
Van Deerlin VM, Sleiman PM, Martinez-Lage M et al (2010) Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet 42(3):234–239
van der Zee J, Gijselinck I, Dillen L et al (2012) A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence. Genomic Instability and intermediate repeats. Hum Mutat. doi:10.1002/humu.22244
Vance C, Rogelj B, Hortobagyi T et al (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323(5918):1208–1211
Watts GD, Wymer J, Kovach MJ et al (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36(4):377–381
Wilm M, Shevchenko A, Houthaeve T et al (1996) Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379(6564):466–469
Wu CH, Fallini C, Ticozzi N et al (2012) Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488(7412):499–503
Zu T, Gibbens B, Doty NS et al (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA 108(1):260–265
Acknowledgments
We thank Iryna Pigur for expert technical assistance, Axel Imhof, Harald Steiner, Akio Fukumori, Richard Page, and Eva Bentmann for providing tools and technologies and Dorothee Dormann for critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB-596), the Competence Network for Neurodegenerative Diseases (KNDD) of the Bundesministerium für Bildung und Forschung (BMBF) to C.H. and the Consortium of Centers of Excellence in Neurodegenerative Brain Diseases (CoEN) to C.H., M.C., D.E., and C.V.B. K.M. was supported by a postdoctoral fellowship from the Alexander von Humboldt Foundation. D.E. was supported by the Helmholtz Young Investigator Program HZ-NG-607. The Agency for Innovation by Science and Technology provides a PhD fellowship to J.J. The authors acknowledge the Antwerp biobank of the Institute Born-Bunge for the brain samples as well as the neurologists S. Engelborghs and P.P. De Deyn and neuropathologist J.J. Martin for the clinical and pathological diagnoses. The Antwerp site is supported for the genetic research of neurodegenerative brain diseases by the Belgian Science Policy Office Interuniversity Attraction Poles program, the Foundation for Alzheimer Research (SAO/FRA), the Medical Foundation Queen Elisabeth, the Flemish Government Methusalem excellence program, the research Foundation Flanders (FWO) and the Special Research Fund of the University of Antwerp, Belgium.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mori, K., Lammich, S., Mackenzie, I.R.A. et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol 125, 413–423 (2013). https://doi.org/10.1007/s00401-013-1088-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-013-1088-7