Transfer learning meets livestock genomics
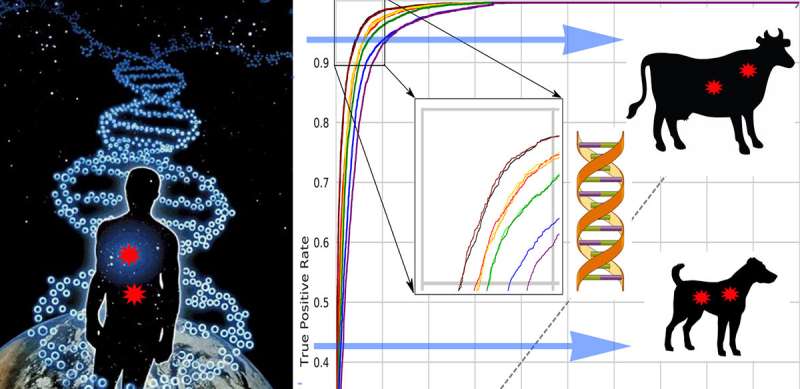
Researchers at Peter the Great St. Petersburg Polytechnic University (SPbPU) have developed a new computational method that predicts harmful mutations in mammalian species. As more livestock producers are using genetic tests to improve their herds, this method will help to optimize and guide the animal breeding programmes, as well as increase the profitability and yields of livestock. Published in Evolutionary Applications, the method follows closely in the footsteps of the most recent innovations in human genomics and translates the knowledge about genetics risk factors in humans to companion animals, thus having a transformative potential for genetics and genomics of livestock species.
Efficient selection in farm animals that will produce offspring with desirable phenotypes and ensure less reliance on hormones and antibiotics has been in the spotlight of many livestock genomic and gene-editing projects. However, their success has been hindered by unknown effects produced by genomic variants. Scientists, led by Profs. Maria Samsonova, Sergey Nuzhdin and Lev Utkin (Mathematical Biology and Bioinformatics Lab and Machine Learning Group) took the full advantage of available information on human variation with deleterious potential and used transfer learning methods to enable classification of damaging mutations in other mammalian species. The approach was extensively validated using dog and mouse models. "The developed methodology could be used to identify deleterious, unwanted mutations in genomes of other farm animals, thus facilitating the design of finely tuned metabolic pathways that allow animals to thrive under a wide range of conditions," said Maria Samsonova, head of the Laboratory of Mathematical Biology and Bioinformatics SPbPU.
From the earliest domestication of animals, humans have selected for desirable characteristics or traits in livestock, and conversely, selection against undesirable phenotypes. A side effect of controlled breeding is rapid accumulation of harmful mutations as genomic variants with negative potential are not counterweighted by an inflow of "good" genes from external populations. Over time, this leads to overall reduction of fitness in cattle and other farm animals. Sustainable agriculture and industry's ability to provide humans with higher quality, healthier and lower-cost farm products heavily relies on the knowledge of functional effects induced by either naturally occurring or technologically introduced mutations and our ability to eliminate harmful ones from future breeds.
More information: Elena Plekhanova et al, Prediction of deleterious mutations in coding regions of mammals with Transfer learning, Evolutionary Applications (2018). DOI: 10.1111/eva.12607
Provided by Peter the Great Saint-Petersburg Polytechnic University

















