Abstract
Background
In utero transmission of SARS coronavirus 2 (SARS-CoV-2) has not been fully investigated. We investigated whether newborns of mothers with COVID-19 during pregnancy might harbor SARS-CoV-2 in the gastrointestinal tract.
Methods
This cohort study investigated stool from 14 newborns born at 25–41 weeks admitted at delivery to our urban academic hospital whose mothers had COVID-19 during pregnancy. Eleven mothers had COVID-19 resolved more than 10 weeks before delivery. Newborn stool was evaluated for SARS-CoV-2 RNA, Spike protein, and induction of inflammatory cytokines interleukin-6 (IL-6) and interferon-γ (IFN-γ) in macrophages.
Results
Despite negative SARS CoV-2 nasal PCRs from all newborns, viral RNAs and Spike protein were detected in the stool of 11 out of 14 newborns as early as the first day of life and increased over time in 6. Stool homogenates from all 14 newborns elicited elevated inflammatory IL-6 and IFN-γ from macrophages. Most newborns were clinically well except for one death from gestational autoimmune liver disease and another who developed necrotizing enterocolitis.
Conclusions
These findings suggest in utero transmission of SARS-CoV-2 and possible persistent intestinal viral reservoirs in the newborns. Further investigation is required to understand the mechanisms and their clinical implications.
Impact
-
SARS-CoV-2 RNAs or Spike protein was detected in the stool of 11 out of 14 preterm newborns born to mothers with resolved COVID-19 weeks prior to delivery despite negative newborn nasal PCR swabs.
-
These novel findings suggest risk of in utero SARS-CoV-2 transmission to the fetal intestine during gestation.
-
The presence of SARS-CoV-2 RNAs and Spike protein in the intestines of newborns may potentially impact the development of the gut microbiome and the immune system; the long-term health impact on the preterm infants should be further investigated.
Similar content being viewed by others
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in pregnancy is associated with adverse pregnancy outcomes, maternal morbidity and mortality, and neonatal complications.1,2 The impact on the fetus remains unclear. However, the overwhelming majority of newborns born to coronavirus disease 2019 (COVID-19)-positive mothers are asymptomatic at delivery, and even when COVID-positive exhibit a benign neonatal course. Initial assessment of the newborn for evidence of COVID-19 is performed via nasal testing using polymerase chain reaction (PCR). Additional workup is performed only when results are positive, including blood cultures, antibody testing, urine and CSF testing, when indicated, to assess for spread of the virus. Over the 16 months since the beginning of the pandemic, over 250 newborns, both term and preterm, have been tested in our institution for COVID-19 using SARS CoV-2 nasal PCR and all results have been negative.3,4
SARS-CoV-2 virus has been found in the nares of infants born to mothers with active COVID-19 at delivery, likely due to horizontal transmission.5,6 Compared to older age groups with SARS-CoV-2 infection, the rates of hospitalization (62%) or admission to intensive care (5%) are higher for children under 1 year of age infected with SARS-CoV-2.7 To date, our understanding of neonates and premature infants infected with SARS-CoV-2 remains limited.
Proinflammatory cytokines play integral roles in the immune response to pathogens. Interleukin-6 (IL-6) is an innate proinflammatory cytokine produced in copious amounts mainly by innate immune cells such as macrophages during the acute phase of infection, but excessive production of IL-6, as reported in severe COVID-19, can be pathologic and contribute to the development of cytokine storms, autoimmunity and chronic inflammation.8,9 Interleukin-1-beta (IL-1β) is a cytokine that plays a role in the pathogenesis of gastrointestinal disease, such as inflammatory bowel disease.9 Interferon-gamma (IFN-γ) is another cytokine involved in the innate and adaptive immune system that is a potent activator of macrophages.10 Both IL-6 and IFN-γ have been shown to be upregulated in cases of viral infection and in cases of viral and bacterial gastroenteritis in children less than 2 years of age.11 Significant elevations in inflammatory markers, including IL-6 and IFN-γ, in cytokine storms play a central role in the immunopathology of COVID-19, influenced by variations in the microbiome.11
In July 2020, we established a biorepository of stool specimens of preterm and term infants in an effort to investigate the gut microbiome in this population. Serendipitously, we observed in initial exploratory studies significantly elevated inflammatory cytokines, IL-6 and IFN-γ, from mouse macrophages after stimulation with stool homogenates as early as day one of life from mostly premature newborns born to mothers with COVID-19 during pregnancy. This observation led to further investigation of whether newborns born to mothers with COVID-19 during pregnancy might harbor SARS-CoV-2 in the gastrointestinal tract.
The objective of this ongoing study was to evaluate for the presence of COVID-19 viral RNAs and Spike protein in stool specimens of term and preterm newborns, from day of life one to 2 months of age during initial hospital admission at birth, born to mothers mostly (11 out of 14) with COVID-19 infection that had been resolved weeks prior to delivery.
Methods
This was a cohort study. The patient population consisted of 14 newborns of gestational age (GA) 25-41 weeks delivered between July 2020 and May 2021, admitted to the neonatal intensive care unit (NICU) and well nursery immediately at delivery at NewYork Presbyterian-Weill Cornell Medicine and whose mothers had COVID-19 during pregnancy (Table 1). Participants were not included if COVID-19 likely occurred prior to conception. Initially, all patients in the NICU and well nursery were approached for consent for the collection of stool specimens as part of our effort to investigate the neonatal gut microbiome. Initial exploratory studies showed that stool homogenates from infants born to mothers with COVID-19 during pregnancy induced higher levels of inflammatory cytokines produced by macrophages. 14 newborns from the enrollment population had a maternal history of COVID-19 infection during pregnancy, and thus were selected for this study’s COVID cohort. From the population of newborns enrolled within the same period, a group of 30 newborns born to mothers without a reported or documented case of COVID-19 during pregnancy were selected based on similar GAs for comparison. They were included as negative controls for detection of viral RNA or protein in stool specimens, and as a comparison group for gastrointestinal and respiratory symptoms (Supplement Table 2). Matching of these controls was limited by availability of newborns of similar gestational ages admitted to the hospital at the time of our enrollment. The timing of maternal COVID-19 infection during pregnancy was verified by positive SARS CoV-2 nasal PCR swab at time of infection (n = 7), or positive COVID-19 serology (n = 10) with timing based on reported history of typical symptoms (4 had both positive nasal PCR swab and serology).
All stool specimens were collected starting from day one of life if possible from each newborn, and weekly thereafter if stool specimens were available, to a maximum of 87 days of life. All stool was collected during the initial hospital admission at birth. Due to various medical conditions, some newborns had decreased frequency of bowel movements and stool specimens were obtained when available. Direct contact between newborns and mothers or support partners varied and ranged from strictly no contact due to maternal active COVID-19, to less contact due to the infant’s medical requirement to be in an isolette in the NICU, to direct contact in a postpartum room with a breastfeeding mother. One newborn (P12) was transferred from another institution in an isolette via ambulance for a higher level of care.
Fresh stool specimens were collected from diapers by trained nurses with sterile tongue depressors and transferred to sterile urine cups, sealed and stored in a 4 °C refrigerator for up to 6 h before stored in a −80 °C freezer.
Chart review of gastrointestinal and early respiratory history
Chart review was performed to analyze the gastrointestinal and early respiratory history of all subjects (Table 1), including evidence of any respiratory support beyond the first day of life, chest x-ray findings suggestive of lung pathologies, and presence of any gastrointestinal issues throughout the course of hospitalization, including abdominal distention, meconium plug syndrome, liver disease, and gastro-esophageal reflux disease.
The comparison newborns were grouped with newborns of mothers with COVID-19 during pregnancy by gestational age group (24–28, 29–32, 33–36, or 37 weeks or later; [Supplement Table 2]). Ideal matching to all factors was limited to newborns available who were enrolled in our study at that time.
The study was reviewed and approved by the NewYork Presbyterian–Weill Cornell Medicine Institutional Review Board. One parent of each participant provided written informed consent prior to collection of stool specimens. All specimens were anonymously coded prior to processing and the Principal Investigator (PI) and laboratory technician were blinded to protected health information.
Statistical analyses
All statistical analyses were performed using Prism 9 (GraphPad Software, San Diego,CA). Normality was determined using the Shapiro–Wilk normality test. Differences between two groups were evaluated using the unpaired t test (for parametric data) or Mann–Whitney test (for nonparametric data), and comparisons of more than 3 groups were evaluated using ordinary one-way analysis of variance followed by Tukey’s correction for multiple comparisons (for parametric data) or Kruskal–Wallis test followed by Dunn’s correction for multiple comparisons (for nonparametric data). Differences of p < 0.05 were considered significant in all statistical analyses. Statistically significant differences are shown with asterisks as follows: *p < 0.05, **p < 0.01, and ***p < 0.001. Each figure shows data for individual animals or biological replicates.
Macrophage cytokine assay
In all, 1 × 105 of bone marrow-derived macrophages (BMDMs) were plated per well in a 24-well plate and cultured overnight before addition of heat-inactivated stool specimen. One hundred micrograms of each stool specimen was resuspended in 1 mL of phosphate-buffered saline (PBS), thoroughly homogenized, and heat-inactivated at 85 °C for 30 min. Ten microliters of the homogenized stool specimen supernatant was added to the BMDM culture per well. Eighteen hours later, the supernatant was removed and the BMDMs were washed with PBS once before extraction of RNA with TRIzol for real-time quantitative reverse transcription (qRT)-PCR for beta actin, IL-6, IFN-γ, and IL-1β expression. Beta-actin was used for normalization and expression levels of IL-6, IFN-γ, and IL-1β were compared relative to the expression of these genes in unstimulated BMDMs.
Spike protein ELISA
For the detection of the Spike protein in stool samples, 96-well plates were coated with recombinant human soluble angiotensin-converting enzyme 2 (sACE2) at a concentration of 150 ng/mL in PBS overnight at 4 °C. 50 mg of stool was suspended in 250 μL PBS, vortexed vigorously for 3–5 min, and then centrifuged at 10,000 rpm for 30 seconds to clarify. Coated plates were blocked for 2 h with PBS containing 1% bovine serum albumin (BSA), and then 100 μL of the supernatant of homogenized stool was added per well and incubated overnight at 4 °C. Plates were then washed 10 times in PBS containing 0.05% Tween-20 and incubated for 2 h at room temperature with mouse anti-spike antibody (clone 1A9; Genetex GTX632604) diluted 1:500 in PBS with 1% BSA. Plates were washed 5 times in PBS with 0.05% Tween-20 and incubated for 2 h at room temperature with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG diluted 1:5000 in PBS with 1% BSA. Plates were washed 5 times in PBS with 0.05% Tween-20 and 100 μL/well of tetramethylbenzidine (TMB) substrate for HRP was added. The colorimetric reaction was allowed to develop for 10 minutes before quenching with 50 μL/well of 2 N sulfuric acid, and absorbance was measured at 450 nm.
Recombinant human sACE2 was generated in CHO-K1 cells (ATCC #CCL-61) transfected with an expression plasmid containing sACE2 (a.a. 1–732) and an 8-histidine tag (Addgene #145101).2 sACE2 was purified out of CHO-K1 cell supernatant using HisPur NiNTA columns (Thermo Fisher Scientific, Waltham, MA) and eluted with a step elution of PBS supplemented with 20, 50, 411, and 250 mM imidazole. The 250 mM imidazole fraction was concentrated with a 30 kD MWCO centrifugal device (MilliporeSigma), and stored at −80 °C until use.
qRT-PCR for viral RNAs in placental tissues
Total RNA was extracted using TRIzol (Thermo Fisher Scientific) followed by ezDNAse treatment (Thermo Fisher Scientific, Waltham, MA) per the manufacturer’s instructions. To quantify viral subgenomic N transcripts, one-step quantitative real-time PCR was performed using the SuperScript III Platinum SYBR Green One-Step qRT–PCR Kit (Invitrogen) with primers, as previously described. Reactions were performed on a QuantStudio 6 Flex Real Time PCR Instrument (Applied Biosystems). The delta-delta-cycle threshold (ΔΔCT) = −(Ct of a stool specimen within COVID-19 cohort − average Ct of non-COVID-19 negative cohort). This means that stools were considered positive for SARS-CoV-2 viral RNA if the Ct value was lower than the average Ct of negative control newborns (newborns born to mothers without reported or documented COVID-19 during pregnancy).
qRT-PCR for stool viral RNAs
Viral RNA was extracted from 100 μg of stool specimen using GeneJet Viral RNA kit (Thermo Fisher Scientific) per the manufacturer’s protocol. In all, 100 ng of RNA was used for RT-PCR to generate complementary DNA (cDNA). One microliter of the cDNA was used per qPCR reaction to quantify viral RNA using the CDC NSP14 and Envelope primers, as indicated below. ΔΔCT values were determined relative to non-COVID samples. Positive controls (cDNAs from infected kidney organoids) were included in each qPCR.
Results
The patient population consisted of 14 newborns of gestational age (GA) 25–41 weeks born to mothers with COVID-19 during pregnancy and 30 comparison newborns of GA 26 2/7–39 6/7 weeks born to mothers with no reported or documented COVID-19. There were 13 mothers studied, including one mother with twins. Asymptomatic mothers (n = 11) had resolved symptoms of COVID-19 on admission to labor and delivery (Table 1) with resolved infection at least 10 weeks prior to delivery. Two mothers with active infection (P10 and the mother of twins P13 and P14) had symptomatic infection at delivery. Based on our hospital’s guidelines as of May 11, 2021, a patient with a positive test obtained between 10 days and 3 months prior to a labor and delivery admission was considered “COVID-recovered” and no repeat SARS CoV-2 nasal PCR testing was required. As such, we considered all but the two mothers with active infection to be fully recovered at delivery and likely unable to infect their newborns via contact or droplet means. Demographics of the study participants are summarized in Table 1.
Ten mothers were tested for COVID-19 IgG serologies against the N antibody or nucleocapsid protein and all results were positive; three mothers were not tested for serologies (Table 1). All newborns were tested with SARS CoV-2 nasal PCR swabs at varying time points between 24 h of life to 2 weeks of life and all results were negative. None of the newborns in the comparison non-COVID cohort were tested with SARS CoV-2 nasal PCR swabs.
Most newborns in this cohort had unremarkable hospitalizations. Many were admitted to the NICU for prematurity or respiratory distress with typical radiographic findings of either transient tachypnea of the newborn or respiratory distress syndrome (Table 1). Notably, two newborns exhibited significant clinical symptoms, outlined below as case 1 and case 2.
Increased stimulation of IL-6 and IFNγ production by stool from COVID cohort
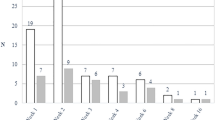
Heat-inactivated homogenates of stool specimens of newborns born to mothers with previous COVID-19 or newborns in the comparison group were co-cultured with macrophages overnight. The expression of IL-6, IFN-γ, and IL-1β were measured by qPCR. The stool homogenates from the COVID cohort induced increased levels of IL-6 and IFN-γ in mouse macrophages (Fig. 1).
Expression of IL-6, IFN-γ, and IL-1β by mouse macrophages after co-culture with heat-inactivated stool specimen for 18 h. Stool specimens from earliest time points (1–2 time points) from the COVID and GA-matched non-COVID groups were used. More than one time point were analyzed for some of the infants who had multiple samples collected. *p < 0.05; 2-way ANOVA.
Detection of SARS-CoV-2 viral RNA and Spike protein in stool specimens
Viral RNAs of the non-structural protein 14 (NSP14) and envelope protein (E) of SARS-CoV-2 were quantitatively detected in the stool from 7 of 14 newborns obtained from 1-87 days after birth (Fig. 2a, c and Fig. 3a). RNA expression was detected by qRT-PCR, where ΔΔCt values indicate RNA expression. RNA levels increased over time in the stool of two newborns (Fig. 3a). The Spike protein, a SARS-CoV-2 surface protein that mediates viral entry, was detected by enzyme-linked immunoassay (ELISA). Spike protein was consistently detected at high levels in 5 newborns (Fig. 2b, c and increased over time in 4 of the newborns (Fig. 3b).
a ∆∆Ct values of stool specimens collected from the COVID (n = 14) and non-COVID (n = 11) groups using the primers for NSP 14. **p < 0.001; unpaired t test. Positive controls were kidney organoids infected with SARS-CoV-2. b Detection of the Spike protein in the stool samples of COVID (n = 14) and non-COVID (n = 30) neonates by ELISA. Relative amounts of Spike in all stool specimens were shown as absorbance at O.D. 450 after subtracting background levels. **p < 0.001; unpaired t test. The stool specimens with the highest viral or Spike protein were included for each neonate; GA-matched samples from neonates without maternal COVID-19 history were used for comparison. c Summary of the timeline of maternal infection, COVID-19 status on mothers and neonates, and detection of SARS-CoV-2 viral RNAs and Spike protein in the stool of each neonate.
a ∆∆Ct values of stool specimens from infants P1, P4, P7, and P10 at different days of life. ∆∆Ct values were calculated with qRT-PCR results using the NSP 14 primers relative to the expression in non-COVID samples. The samples analyzed were all the samples collected from all four infants. Sample availability was dependent on the timing of enrollment, duration of hospitalization, and stool frequency of each infants. b Amounts of the Spike protein in stool specimens of COVID infants P1, P2, P6, P7, and P9 at different days of life.
Case 1
One neonate (P4) was born at 780 grams and 30 1/7 weeks GA to a mother with mild upper respiratory symptoms of COVID-19 infection 5 months prior to delivery; her symptoms resolved well before delivery. Upon admission to labor and delivery, the asymptomatic mother tested positive for nasal viral RNA and serum viral IgG antibodies. P4 had multiple negative nasal COVID-19 PCR swabs and positive COVID-19 serum IgG antibodies. He presented immediately after birth in severe and persistent liver failure and died at age 11 weeks due to complications of liver failure. Autopsy findings were compatible with his clinical diagnosis of gestational alloimmune liver disease (GALD) with a phenotype of neonatal hemochromotomasis. Stool specimens of P4 showed increasing levels of viral RNA up until day 56 (Fig. 3a).
Case 2
Another newborn (P6) developed necrotizing enterocolitis (NEC). P6 is a 27 4/7 week GA female whose mother had a positive COVID-19 PCR test and mild viral upper respiratory symptoms at approximately 15 weeks of gestation with symptoms that also resolved long before delivery. P6 initially had an uncomplicated medical course in the first few weeks of life. She developed NEC on day of life 26 at 31 weeks corrected gestational age on fortified full enteral feedings. She developed abdominal compartment syndrome quickly requiring a bedside abdominal decompression and jejunal and ileal resections. Increasing levels of the Spike protein were found in her stool specimens from day of life 6 to 15, suggestive of active viral replication (Fig. 3b).
Discussion
This is an initial report describing the presence of SARS-CoV-2 RNA and protein in the stool of 14 preterm newborns during hospital admission at delivery with negative SARS CoV-2 nasal PCR results and born to mothers with COVID-19 infection during pregnancy. The Spike protein, a SARS-CoV-2 surface protein that mediates viral entry, was consistently detected at high levels in one-third of infants.
Notably, maternal COVID-19 infection in our study was at least 10 weeks remote from delivery (Fig. 2c and Table 1), and 11 out of 13 mothers in this cohort had no active COVID-19 symptoms at delivery, making direct contact, droplet, or airborne transmission unlikely. Stool was collected as early as the first day of life. This suggests in utero SARS-CoV-2 transmission to the fetal intestine as a mechanism for the virus to be transmitted to the newborns. Preterm twins (P13 and P14), born to a mother who delivered in the adult intensive care unit due to severe COVID-19, showed no detectable viral RNA or protein in their stool (Fig. 2c and Table 1). We speculate that in utero transmission in this case may have been unlikely at 31 weeks GA due to more developed mechanisms to protect the fetus. Earlier pregnancy is associated with higher placental expression of viral entry receptors, including angiotensin-converting enzyme 212 and neuropilin.13 This suggests that in utero transmission of SARS-CoV-2, if possible, may be more likely during earlier gestion.
The mechanism of transmission to the fetal intestine remains unclear. In utero transmission of viruses, such as CMV, is transplacental. Additionally, SARS-CoV-2 has been detected in placentas.13,14 Viral RNAs were detected in the placentas of two mothers in this cohort with active COVID-19 at delivery (Table 1). SARS-CoV-2 viral RNAs have also been detected in amniotic fluid,15 which may be ingested by the fetus and passed through to the fetal intestine in utero.
Currently, our understanding of the impact of antenatal COVID-19 on the newborn is limited. SARS CoV-2 nasal PCRs performed in the immediate post-natal period of this cohort of newborns born to mothers with COVID-19 during gestation at our institution have been negative at the time of data analysis.16 However, negative nasal PCRs do not exclude the possibility of SARS-CoV-2 present in other tissues of the newborn, which have not been rigorously studied. A recent study by Boateng et al. reported no detection of SARS-CoV-2 viral RNA in the stool and urine from term newborns whose mothers had COVID-19 during pregnancy.17 However, the newborns in this study by Boateng et al. were term or near term (average GA ~39 weeks), compared to the average GA of 34 weeks in our NICU cohort. Our findings suggest increased risk of in utero transmission when maternal COVID-19 occurred prior to ~27 weeks of gestation. Furthermore, all stool and urine samples were collected at 15 h following birth in the study by Boateng et al.; viral titers might be below the detection limit at such an early time. In contrast, many of our samples were tested at multiple time points up to two months of age. Therefore, there are inherent differences in the infant cohorts and timing of fecal sample collection between our study and the study by Boateng et al.
Another study by Zeng et al.5 describes 33 neonates, including 30 with negative nasopharyngeal and anal PCR swabs born to mothers with infection at various time points during pregnancy. 3 neonates were diagnosed with active COVID-19 with positive nasopharyngeal and anal swabs, and all 3 were born to mothers diagnosed with active infection at delivery. The negative anal swabs described may only have reflected the lack of RNA present on the skin’s surface and may have been taken prior to any stooling. In contrast, our study directly assessed the presence of both SARS-CoV-2 viral RNA and protein in newborn stool reflective of intestinal lumen contents. The majority of our infants (11 of 14) were born to mothers with COVID-19 resolved more than 10 weeks prior to delivery; all of these 11 infants had SARS-CoV-2 RNA and Spike protein detected in the stool.
Persistent viral reservoirs in the intestine have been found in both recovered adult and pediatric COVID-19 patients 4 months after infection, suggesting the intestinal environment providing a unique niche for prolonged viral reservoirs.18,19 Enterocytes and smooth muscle cells in the intestine express high levels of ACE2 and neuropilin, respectively.20,21 A recent study demonstrates high expression of ACE2 in the human fetal intestine in early second trimester.22 Additionally, the intestinal lumen is less accessible to immune cells. These factors may collectively create a niche in the intestinal lumen that is conducive to the establishment of what may be persistent viral reservoirs of SARS-CoV-2 in the fetus and newborn.
The clinical implications of our findings of viral RNAs and Spike protein in the stool of these newborns require further investigation. Most newborns studied here were either clinically well or progressed as expected during hospital admission for their gestational ages. There were two symptomatic cases presented in this report. While there is insufficient evidence to connect the development of symptoms to effects of COVID-19, It remains unclear if the presence of intestinal reservoirs of SARS-CoV-2 might have contributed to the development of GALD or NEC in these two newborns. Maternal viral infections known to cause maternal immune activation, including herpes simplex virus, enterovirus and cytomegalovirus, have been associated with the development of acute liver failure in newborns, which is commonly caused by GALD.23 The immunopathology of COVID-19 involves excessive inflammatory response, impaired adaptive immunity, and production of autoantibodies.24,25,26 Infections during pregnancy may increase the risk of maternal alloimmune responses, such as GALD, whereby maternal IgG antibodies attack fetal hepatocyte antigens, resulting in fetal and neonatal liver injury. Autoantibodies that antagonize type I IFN responses were found in 19% of patients with critical COVID-19.27 Autoantibody reactivities in pregnant women with COVID-19 may present potential additional risk to the fetus, as maternal autoantibodies unleashed by SARS-CoV-2 infection may attack fetal cells. Further investigation is required to elucidate autoantibody responses in pregnant women with COVID-19 and possible risk to the fetus.
The case of NEC raises the concern that intestinal inflammation driven by an intestinal reservoir of SARS-CoV-2 may increase the risk of gastrointestinal complications in these newborns, such as NEC. COVID-19 has previously been implicated in the development of NEC in newborns.28,29 However, NEC may have developed irrespective of maternal COVID-19 status in this newborn who was already at high risk due to prematurity.
IL-6 and IFN-γ are both proinflammatory cytokines that have been reported to be elevated in severe COVID-19 patients.30,31 In particular, as a potent innate cytokine, IL-6 is thought to contribute to potentially fatal cytokine storms in COVID-19.30,32 Increased induction of these cytokines in the neonatal intestine by viral RNAs may impact the immune cell development and immune landscape in the neonatal intestine and may potentially affect disease susceptibility in later life.
Limitations of this study include a small sample size. Control group newborns born to mothers without a confirmed or reported case of COVID-19 during pregnancy were limited to newborns admitted at the time of our study. Furthermore, the current understanding is that persistent intestinal reservoirs of SARS-CoV-2 may not reflect an active infection.33
It is unclear if the presence of viral RNA and protein within the gut microbiome represents active virus in newborns with clinical hospital courses typical of their gestational age in 12 out of 14 cases. However, increasing levels of viral RNA and protein over time suggest replication in some infants. The stool homogenates from the COVID-19 cohort induced a significant increase in IL-6 and IFN-γ production in macrophages, reflecting higher amounts of inflammatory components in the intestinal lumen of these infants, consistent with the detection of SARS-CoV-2 viral RNA and protein in their stool specimens. Early development of the immune system is heavily influenced by the gut microbiome;34 the full impact of persistent viral reservoirs on the development of the immune system in these infants may not be fully appreciated until years later. Further investigation is required to understand the mechanisms of in utero transmission of SARS-CoV-2 in women with COVID-19 during early stages of gestation, as well as the full impact of persistent intestinal viral reservoirs on child health and development.
Data availability
All data generated or analyzed during this study are included in this article and can be requested from the corresponding author upon request.
References
Villar, J. et al. Maternal and neonatal morbidity and mortality among pregnant women with and without COVID-19 infection: the INTERCOVID Multinational Cohort Study. JAMA Pediatr. 175, 817–826 (2021).
Allotey, J. et al. Clinical manifestations, risk factors, and maternal and perinatal outcomes of coronavirus disease 2019 in pregnancy: living systematic review and meta-analysis. BMJ 370, m3320 (2020).
Perlman, J., Oxford, C., Chang, C., Salvatore, C. & Di Pace, J. Delivery room preparedness and early neonatal outcomes during COVID-19 pandemic in New York City. Pediatrics https://doi.org/10.1542/peds.2020-1567 (2020).
Salvatore, C. M. et al. Neonatal management and outcomes during the COVID-19 pandemic: an observation cohort study. Lancet Child Adolesc. Health 4, 721–727 (2020).
Zeng, L. et al. Neonatal early-onset infection with SARS-CoV-2 in 33 neonates born to mothers with COVID-19 in Wuhan, China. JAMA Pediatr. 174, 722–725 (2020).
Cook, J. et al. Horizontal transmission of severe acute respiratory syndrome coronavirus 2 to a premature infant: multiple organ injury and association with markers of inflammation. Lancet Child Adolesc. Health 4, 548–551 (2020).
CDC COVID-19 Response Team. Coronavirus disease 2019 in children - United States, February 12-April 2, 2020. MMWR Morb. Mortal. Wkly Rep. 69, 422–426 (2020).
Velazquez-Salinas, L., Verdugo-Rodriguez, A., Rodriguez, L. L. & Borca, M. V. The role of interleukin 6 during viral infections. Front. Microbiol. 10, 1057 (2019).
Jones, S. A. & Hunter, C. A. Is IL-6 a key cytokine target for therapy in COVID-19. Nat. Rev. Immunol. 21, 337–339 (2021).
Katze, M. G., He, Y. & Gale, M. Jr Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2, 675–687 (2002).
Chou, J., Thomas, P. G. & Randolph, A. G. Immunology of SARS-CoV-2 infection in children. Nat. Immunol. 23, 177–185 (2022).
Li, M., Chen, L., Zhang, J., Xiong, C. & Li, X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS ONE 15, e0230295 (2020).
Argueta, L. B. et al. SARS-CoV-2 infects syncytiotrophoblast and activates inflammatory responses in the placenta. Preprint at bioRxiv https://doi.org/10.1101/2021.06.01.446676 (2021).
Cribiu, F. M. et al. Severe SARS-CoV-2 placenta infection can impact neonatal outcome in the absence of vertical transmission. J. Clin. Investig. https://doi.org/10.1172/JCI145427 (2021).
Bahadur, G. et al. Retrospective observational RT-PCR analyses on 688 babies born to 843 SARS-CoV-2 positive mothers, placental analyses and diagnostic analyses limitations suggest vertical transmission is possible. Facts Views Vis. Obgyn. 13, 53–66 (2021).
Flannery, D. D. et al. Assessment of maternal and neonatal cord blood SARS-CoV-2 antibodies and placental transfer ratios. JAMA Pediatr. 175, 594–600 (2021).
Boateng, J. O. et al. SARS-CoV-2 in infant urine and fecal samples after in utero COVID-19 exposure. Pediatr. Res. https://doi.org/10.1038/s41390-021-01822-x (2021).
Park, S. K. et al. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 19, 1387–1394.e1382 (2021).
Zhang, T. et al. Detectable SARS-CoV-2 viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J. Med. Virol. 92, 909–914 (2020).
Yamaji, M., Mahmoud, M., Evans, I. M. & Zachary, I. C. Neuropilin 1 is essential for gastrointestinal smooth muscle contractility and motility in aged mice. PLoS ONE 10, e0115563 (2015).
Hashimoto, T. et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487, 477–481 (2012).
Elmentaite, R. et al. Cells of the human intestinal tract mapped across space and time. Nature 597, 250–255 (2021).
Ibrahim, S. H. et al. Liver diseases in the perinatal period: interactions between mother and infant. Hepatology 71, 1474–1485 (2020).
Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science https://doi.org/10.1126/science.abd4585 (2020).
Zuo, Y. et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.abd3876 (2020).
Chang, S. E. et al. New-onset IgG autoantibodies in hospitalized patients with COVID-19. Nat. Commun.12, 5417 (2021).
van der Wijst, M. G. P. et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.abh2624 (2021).
Wu, Y. T. et al. Neonatal outcome in 29 pregnant women with COVID-19: A retrospective study in Wuhan, China. PLoS Med. 17, e1003195 (2020).
Mehl, S. C. et al. Necrotizing enterocolitis-like pneumatosis intestinalis in an infant with COVID-19. Pediatr. Infect. Dis. J. 40, e85–e86 (2021).
Investigators, R.-C. et al. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N. Engl. J. Med. 384, 1491–1502 (2021).
Heuberger, J. et al. Epithelial response to IFN-gamma promotes SARS-CoV-2 infection. EMBO Mol. Med. 13, e13191 (2021).
Chen, L. Y. C., Hoiland, R. L., Stukas, S., Wellington, C. L. & Sekhon, M. S. Assessing the importance of interleukin-6 in COVID-19. Lancet Respir. Med. 9, e13 (2021).
Gaebler, C. et al. Evolution of antibody immunity to SARS-CoV-2. Nature 591, 639–644 (2021).
Sanidad, K. Z. & Zeng, M. Y. Neonatal gut microbiome and immunity. Curr. Opin. Microbiol. 56, 30–37 (2020).
Acknowledgements
We would like to thank all participants in this research study for their continued support of neonatal microbiome research.
Funding
This work was supported by NIH grant 5 K01 DK114376 and funds from the Gale and Ira Drukier Institute for Children’s Health and Children’s Health Council at Weill Cornell Medicine, the Center for Immunology and Office of Academic Integration of Cornell University, the Center for IBD Research at Weill Cornell Medicine, and the Hartwell Foundation (all to M.Y.Z.), a Hartwell Foundation Postdoctoral Fellowship (to K.Z.S.), a CTSC TL1 Scholar Award at Weill Cornell Medicine and the Biocodex Microbiota Foundation Grant (to J.A.B.) and the NCI R01CA234614, NIAID 2R01AI107301, and NIDDK R01DK121072 and 1RO3DK117252, Department of Medicine, Weill Cornell Medicine (to R.E.S.), and a Weill Cornell Medicine COVID-19 Research Grant (H.S., R.E.S., R.N.B.). R.E.S. is an Irma Hirschl Trust Research Award Scholar.
Author information
Authors and Affiliations
Contributions
Each author has met the Pediatric Research authorship requirements. M.Y.Z. conceived and supervised the study, analyzed data, and wrote the manuscript. J.C.J. collected stool specimens, performed experiments, analyzed data, and wrote the manuscript. J.M.P. helped with specimen collection and advised the study. A.A. and J.A.B. performed experiments and analyzed data. S.L.R. collected stool specimens and analyzed data. Y.B., K.Z.S., and M.A. assisted with experiments. R.N.B., H.S., and R.E. helped with analysis of placental tissues. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.Y.Z. is a consultant for Guidepoint. R.E.S. is on the scientific advisory board of Miromatrix Inc. and is a consultant and speaker for Alnylam Inc. The rest of the authors have no conflicts of interest to disclose.
Ethics approval and consent to participate
Parents of all subjects gave informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jin, J.C., Ananthanarayanan, A., Brown, J.A. et al. SARS CoV-2 detected in neonatal stool remote from maternal COVID-19 during pregnancy. Pediatr Res 93, 1375–1382 (2023). https://doi.org/10.1038/s41390-022-02266-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-022-02266-7
This article is cited by
-
SARS-CoV-2 reservoir in post-acute sequelae of COVID-19 (PASC)
Nature Immunology (2023)