Although gut viromes are unique from person to person, like fingerprints, they also change as people age. Specifically, gut viromes tend to become more diverse as people reach adulthood, and less diverse as they reach their elder years.
Age-dependent diversity changes in the gut virome parallel age-dependent diversity changes in gut bacterial microbiome, except that in infants with underdeveloped immune systems, gut microbiomes are teeming with a range of virus types, but with relatively few bacteria types.
The new findings are just the first to come from the Gut Virome Database (GVD), which has been compiled by scientists at Ohio State University. The GVD incorporates data from many smaller studies and benefits from machine learning technology to overcome the limitations of existing reference databases. Offering unprecedented scale and richness, the GVD can help scientists survey the sequence space of viruses, which remains largely unexplored.
In an article that appeared August 24 in Cell Host & Microbe, the Ohio State scientists wrote: “Here, we (1) built a human GVD from 2,697 viral particle or microbial metagenomes from 1,986 individuals representing 16 countries, (2) assess its effectiveness, and (3) report a meta-analysis that reveals age-dependent patterns across healthy Westerners. The GVD contains 33,242 unique viral populations (approximately species-level taxa) and improves average viral detection rates over viral RefSeq and IMG/VR nearly 182-fold and 2.6-fold, respectively.”
The GVD’s superior viral detection rates could help scientists navigate the “dark matter” of the gut virome, which complicates metagenomic study. Gut bacteria possess universal phylogenetic markers; gut viruses, however, lack such genetic commonalities.
“We’ve established a robust starting point to see what the virome looks like in humans,” said study co-author Olivier Zablocki, a postdoctoral researcher in microbiology at Ohio State. “If we can characterize the viruses that are keeping us healthy, we might be able to harness that information to design future therapeutics for pathogens that can’t otherwise be treated with drugs.”
The researchers plan to update the GVD, open-access database, on a regular basis. They see the GVD as a “compact, foundational resource” and a set of “standardization guidelines” that can “help maximize our understanding of viral roles in health and disease.” Already, the researchers say, the GVD has been used to confirm findings from previous studies while identifying new trends.
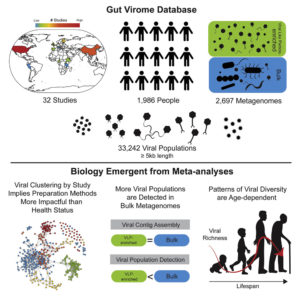
Besides corroborating earlier findings, the GVD uncovered viral diversity changes from human infancy into senescence. The GVD also indicated that people living in non-Western countries have higher gut virus diversity than Westerners.
According to lead author Ann Gregory, who contributed to the study while she was a graduate student at Ohio State, previous research had shown that non-Western individuals who move to the United States or another Western country lose microbiome diversity, suggesting diet and environment drive virome differences.
Variations in viral diversity could also be seen in healthy versus sick participants. “A general rule of thumb for ecology is that higher diversity leads to a healthier ecosystem,” Gregory noted. “We know that more diversity of viruses and microbes is usually associated with a healthier individual. And we saw that healthier individuals tend to have a higher diversity of viruses, indicating that these viruses may be potentially doing something positive and having a beneficial role.”
The most-studied viruses kill their host cells, but scientists in the Ohio State lab in which Gregory and Zablocki worked have discovered more and more phage-type viruses that coexist with their host microbes and even produce genes that help the host cells compete and survive. In fact, almost 97.7% of the virus populations they studied consisted of phages, which are viruses that infect bacteria.
The leader of that lab, senior study author Matthew Sullivan, has his sights set on “phage therapy”—the 100-year-old idea of using phages to kill antibiotic-resistant pathogens or superbugs.
“Phages are part of a vast interconnected network of organisms that live with us and on us, and when broad-spectrum antibiotics are used to fight against infection, they also harm our natural microbiome,” Sullivan said. “We are building out a toolkit to scale our understanding and capabilities to use phages to tune disturbed microbiomes back toward a healthy state.
“Importantly, such a therapeutic should impact not only our human microbiome, but also that in other animals, plants, and engineered systems to fight pathogens and superbugs. They could also provide a foundation for something we might have to consider in the world’s oceans to combat climate change.”