Today's issue of Nature contains a paper with a rather unusual author list. Read past the standard collection of academics, and the final author credited is... an online gaming community.
Scientists have turned to games for a variety of reasons, having studied virtual epidemics and tracked online communities and behavior, or simply used games to drum up excitement for the science. But this may be the first time that the gamers played an active role in producing the results, having solved problems in protein structure through the Foldit game.
According to a news feature on Foldit, the project arose from an earlier distributed computing effort called Rosetta@home. That project used what has become the standard approach for home-based scientific work: a screensaver that provided a graphical frontend to a program that uses spare processor time to solve weighty scientific problems. For Rosetta, that problem was the task of figuring out how proteins, which are composed of a chain of chemicals called amino acids, adopt their final, three-dimensional shape.
This is typically an energy minimization problem. Proteins tend to form structures that keep hydrophobic parts buried internally, away from the water they're dissolved in. They also form bridges between neighboring sections by hydrogen bonds and charge interactions. Maximize these sorts of interactions and you minimize the energy involved.
It sounds simple, but with anything more than a short chain of amino acids, there are a tremendous number of potential configurations to be sampled in 3D space, which can bring powerful computers to their knees.
The Rosetta algorithm handles the huge energy landscape it needs to scan by taking big leaps between different configurations, then attempting to minimize the energy by making smaller tweaks. This lets it sample large portions of the structural landscape, but sometimes leaves it stuck: the path between its current location and an energy minimum may take it through a high energy state, which would keep Rosetta from finding the solution.
Apparently, the program's home users noticed that the screensaver would often show the program stuck close to a much better structure. One of Foldit's developers is quoted as saying, "People started writing in, saying, 'I can see where it would fit better this way.'"
The Rosetta team decided to give them a chance to see if they really could.
Starting with algorithms, ending with brains
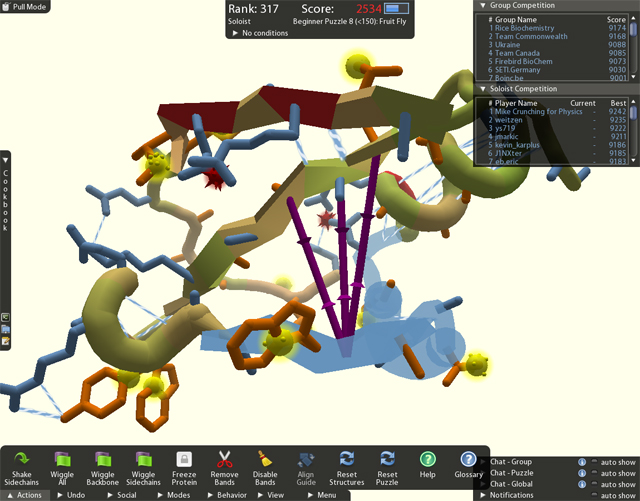
Foldit takes a hybrid approach. The Rosetta algorithm is used to create some potential starting structures, but users are then given a set of controls that let them poke and prod the protein's structure in three dimensions; displays provide live feedback on the energy of a configuration.
Foldit uses some of the same conventions typical of other computer games, like a few simple structural problems to give new users a smooth learning curve. It also borrows from other online gaming communities; there are leaderboards, team and individual challenges, user forums, and so on.
Though very few of those who played Foldit had any significant background in biochemistry, the gamers tended to beat Rosetta when it came to solving structures. In a series of ten challenges, they outperformed the algorithms on five and drew even on another three.
By tracing the actions of the best players, the authors were able to figure out how the humans' excellent pattern recognition abilities gave them an edge over the computer. For example, people were very good about detecting a hydrophobic amino acid when it stuck out from the protein's surface, instead of being buried internally, and they were willing to rearrange the structure's internals in order to tuck the offending amino acid back inside. Those sorts of extensive rearrangements were beyond Rosetta's abilities, since the energy changes involved in the transitions are so large.
Similarly, Rosetta was good at linking up stretches of protein through charge interactions and hydrogen bonds, but it would often get things slightly off (think of a zipper that's off by a single tooth). Shifting every bond by a single partner was beyond Rosetta's abilities, but it's something a human can do trivially.
That's not to say the Rosetta algorithm didn't play a valuable role in Foldit. Humans turn out to be really bad at starting from a simple linear chain of amino acids; they need a rough idea of what the protein might look like before they can recognize patterns to optimize. Given a set of 10 potential structures produced by Rosetta, however, the best players were very adept at picking the one closest to the optimal configuration.
The authors also note that different players tended to have different strengths. Some were better at making the big adjustments needed to get near an energy minimum, while others enjoyed the fine-scale tweaking needed to fully optimize the structure. That's where Foldit's ability to enable team competitions, where different team members could handle the parts of the task most suited to their interests and abilities, really paid off.
The Nature article makes it clear that researchers in other fields, including astronomy, are starting to try similar approaches to getting the public to contribute something other than spare processor time to scientific research. As long as the human brain continues to outperform computers on some tasks, researchers who can harness these differences should get a big jump in performance.
Nature, 2010. DOI: 10.1038/nature09304 (About DOIs).
Listing image by Foldit team, University of Washington.
reader comments
67