For the first time, the structure of a key player in amyotrophic lateral sclerosis (ALS), and multiple other neurodegenerative diseases, has been determined. The abnormal clumping of the TAR DNA-binding protein 43 kDa (TDP-43) in nerve cells is the defining pathological hallmark of ALS. This discovery could enable targeted development of new medical interventions and diagnostic tests, a breakthrough given that no disease-modifying therapies exist for these conditions and early diagnosis is not possible.
This work is published in Nature, in the paper, “Structure of pathological TDP-43 filaments from ALS with FTLD.”
TDP-43 is found in healthy cells throughout the body, but it clumps together forming ‘aggregates’ in the brains of ALS patients. Although scientists have been aware of this for some time, the potential to translate this information into treatments has been limited as the molecular structure of the TDP-43 aggregates has been unknown.
Now, researchers have used cryo-electron microscopy (cryo-EM) to determine the structures of aggregated TDP-43 in the frontal and motor cortices of an individual who had ALS with FTLD and from the frontal cortex of a second individual with the same diagnosis.
The structure of TDP-43 observed in this study was consistent in samples from different regions of the brains of both individuals, but was different from that seen in previous studies that attempted to recreate TDP-43 aggregates in a test tube.
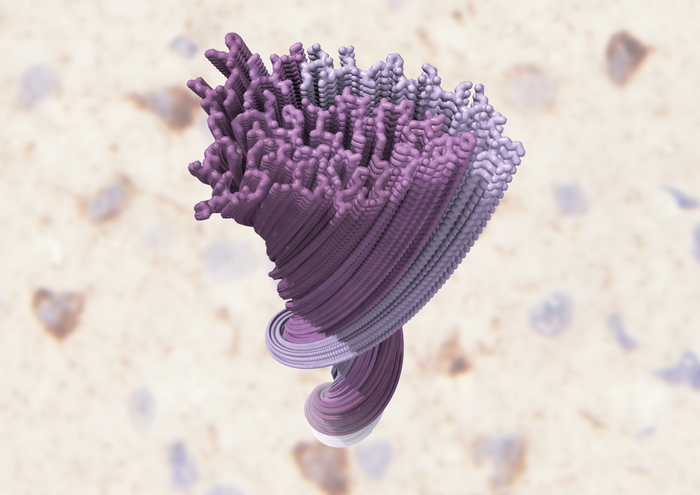
The findings uncovered previously unseen structural features such as a filamentous double-spiral-shaped fold. More specifically, the authors note that an amyloid-like filament structure comprising a single protofilament was found in both brain regions and individuals. The ordered filament core, they write, “spans residues 282–360 in the TDP-43 low-complexity domain and adopts a previously undescribed double-spiral-shaped fold, which shows no similarity to those of TDP-43 filaments formed in vitro. An abundance of glycine and neutral polar residues facilitates numerous turns and restricts β-strand length, which results in an absence of β-sheet stacking that is associated with cross-β amyloid structure.”
It was previously thought that TDP-43 interacted similarly to analogous proteins associated with other neurodegenerative diseases such as Alzheimer’s disease. This study suggests however that aggregation of TDP-43 likely results in different disease mechanisms.
These distinct structural features mean that TDP-43 in the brain is likely to interact uniquely with diagnostic tools and drugs. These differences could explain why current diagnostic compounds based on analogous proteins associated with other neurodegenerative diseases are poor at diagnosing ALS.
The study presents new possibilities for the development of treatments using compounds that specifically target structural features of TDP-43, including those responsible for aggregation.
“There are no diagnostics or therapeutics for ALS and other diseases associated with TDP-43 and the first step towards developing these is gaining a better understanding of TDP-43 itself,” notes Benjamin Ryskeldi-Falcon, PhD, group leader at MRC Laboratory for Molecular Biology in Cambridge, U.K. “Now that we know what the structure of aggregated TDP-43 looks like and what makes it unique, we can use it to find better ways to diagnose the disease early. We’re excited to be able to use this blueprint in our lab to start identifying compounds that bind to unique sites on TDP-43, with the aim of identifying potential therapies for further study.”
“I would especially like to thank the people with ALS, and their families, who donated their brains to research to help us gain a better understanding of this terrible disease,” Ryskeldi-Falcon adds.